
What we do
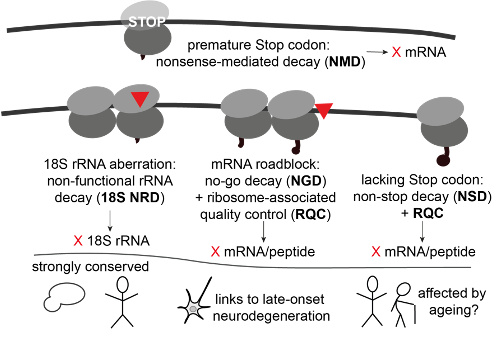 Proteins are central to any living organism. To protect cells from dysfunctional and toxic, disease-causing proteins formed by aberrant translation, eukaryotic co-translational quality control pathways detect translation defects and remove aberrant protein products, together with associated faulty mRNA blueprints and sometimes faulty ribosomal components. Our team is mostly interested in what happens when ribosomes get stuck during translation, and what happens when following ribosomes collide into them. To respond to these molecular 'car crashes', cells have evolved various molecular 'emergency services'. For an overview of what is known so far in the field, have a look at our review Molecular Highway Patrol for Ribosome Collisions. While this is a fast-developing field, much is still unknown about the molecular mechanisms underlying translation quality control, and how these interact with other cellular machineries and with organismal health and life cycle. These are research objects in my group. We use sequencing-based methods, for example UV crosslinking and analysis of cDNA (CRAC), and nanopore sequencing, as well as different molecular biology and biochemical methods to study RNA-protein interactions, translation and RNA modification in the context of translation quality control. These techniques provide a unique toolbox to approach our key aims:
Proteins are central to any living organism. To protect cells from dysfunctional and toxic, disease-causing proteins formed by aberrant translation, eukaryotic co-translational quality control pathways detect translation defects and remove aberrant protein products, together with associated faulty mRNA blueprints and sometimes faulty ribosomal components. Our team is mostly interested in what happens when ribosomes get stuck during translation, and what happens when following ribosomes collide into them. To respond to these molecular 'car crashes', cells have evolved various molecular 'emergency services'. For an overview of what is known so far in the field, have a look at our review Molecular Highway Patrol for Ribosome Collisions. While this is a fast-developing field, much is still unknown about the molecular mechanisms underlying translation quality control, and how these interact with other cellular machineries and with organismal health and life cycle. These are research objects in my group. We use sequencing-based methods, for example UV crosslinking and analysis of cDNA (CRAC), and nanopore sequencing, as well as different molecular biology and biochemical methods to study RNA-protein interactions, translation and RNA modification in the context of translation quality control. These techniques provide a unique toolbox to approach our key aims:

1: Identify and characterize new players in translation quality control pathways.
2: Study links between RNA modification and ribosome collisions, as well as translation quality control.
3: Characterize mitochondrial effects of translation quality control.
As a longterm aim, with our RNA-centred research, we want to make fundamental contributions to the understanding of co-translational quality control and its links to age-related neurodegenerative diseases, to ultimately inform efforts to cure, prevent or delay the onset of such diseases.