
What We Do: Decoding the Ribosome’s Emergency Response System
- Proteins are life's workhorses, and ribosomes are the machines that synthesize them. Although ribosomal translation is tightly regulated, errors still occur. Aberrant proteins can be dysfunctional and toxic and cause dis
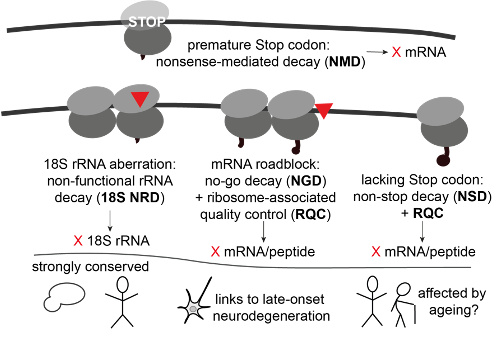 ease.
ease. - To protect themselves, eukaryotic translation quality control pathways detect translation defects and remove aberrant protein products, and associated faulty mRNA blueprints, and sometimes faulty ribosomal components, too.
- We study ribosomes, what happens when they get stuck and following ribosomes collide. and the molecular and cellular 'emergency services' that respond to these molecular 'car crashes'.
- Our review Molecular Highway Patrol for Ribosome Collisions gives an overview of what is known so far. However, much is still unknown about molecular mechanisms underlying translation quality control, and how these interact with other cellular machineries and with organismal health and life cycle.
KEY AIMS - from Molecular Mechanisms to Organismal Implications
We tackle ribosome biology and translation quality control at three levels — molecular, cellular, and organismal. Our aims are to:

(1) Identify and characterize new players in translation quality control pathways.
(2a) Study links between ribosome collisions/translation quality control, ribosome biogenesis & RNA modification.
(2b) Characterize mitochondrial effects of translation quality control.
(3 - long-term) Study links to aging & disease; inform and generate new drugs.
OUR TOOLBOX - Cutting-Edge Methods to Crack the Code
- We use a combination of Molecular Biology, Biochemistry, and cutting-edge sequencing-based methods to study RNA-protein interactions, translation and RNA modification in the context of translation quality control.
- UV crosslinking and analysis of cDNA (CRAC)
- Nanopore sequencing
- These techniques provide a unique toolbox to approach our research aims.
RECENT SUCCESSES - Meet the New Molecular Heroes
- New1 – a protective factor for colliding ribosomes
We identified the fungal protein New1 as a safeguard that prevents ribosome‑collision‑induced no‑go decay of specific mRNAs during termination. Loss of New1 destabilises mRNAs encoding essential metabolic enzymes and translation factors and no-go decay is a major driver of cold sensitivity in New1 mutants.
Methodological highlight: The discovery relied on successful adaptation of nanopore sequencing to identify and quantify no-go decay cleavage events.
Curious? Read our article! - Jlp2 – a novel release factor for stalled peptidyl‑tRNAs
Jlp2 cleaves peptidyl‑tRNA bonds when translation aborts before canonical termination, providing a backup rescue mechanism when ubiquitin‑mediated degradation fails. This way, Jlp2 frees the large ribosomal subunit for reuse and expands the repertoire of translation quality control enzymes.
Want to know more? Read our preprint!
LONGTERM AIM - from Mechanisms to Medicines
Our ultimate goal is to provide a fundamental, mechanistic understanding of ribosome behavior and translation quality control and to translate these insights into strategies for preventing or delaying age‑related neurodegenerative disorders. By establishing links between molecular surveillance pathways and disease phenotypes, we aim to contribute to the design of next‑generation therapeutics.
EAGER to Join the Mission?
We’re always looking for curious, driven students - from B.Sc. to PhD level - who want to tackle the big questions in translation biology.
 ease.
ease.